The first day (Thursday the 19th), there were a number of workshops and tutorials. These included "The Evolution of Physical Systems", "Neuroevolution", "Artificial Life in Industry", and several on AVIDA software application and design. I ran a workshop with Laura Grabowski called the "Hard-to-define Events" (HTDE) workshop. Details are on the slides below.
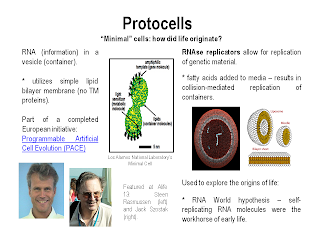
There were a number of talks (including the keynote by Nobel laureate Jack Szostak) on protocells. Protocells are "minimal" cells, or cells that contain the minimal components necessary to enable self-replication, information storage, and metabolic function. Protocells are used to study the origins of life, but also may have many applications in the emerging area of synthetic biology.
There were also a number of papers on collective behavior (or swarm intelligence) in robots and animals. One talk (by Radhika Nagpal) was on using termites as a template for designing robot swarms that can collectively build structures and enable distributed autonomous networks. There were a number of other talks and posters on this topic, mostly focusing on the assembly and control of robot teams.
The simulation of biology (and biological principles themselves) can now be transferred to physical 3-dimensional models. Related to this are models that capture the physical nature of biological systems. To transfer what we learn on a computer to the physical world, CNC machines can be used to "print" these models using a variety of materials. This allows us to test designs (such as robot body plans) using a set of actual physical forces and conditions. To actually model the physical aspects of biology, our toolkit now goes beyond physics engines. One example of this from the conference is the use of model genetic regulatory networks (GRNs) to approximate the emergence of a phenotype in development. See slide below for more details.
A less prominent but still important part of the conference was the art and philosophy track. The art and philosophy behind Artificial Life focuses on what it means to create artificial and engineered life. One speaker (Oron Catts) showed demonstrations of his art involving "semi-living" systems (see slide below for more details).
Much of this conference was focused on discovering and exploring ways to simulate processes associated with evolution, whether it be on a computer, in a petri dish, or in some other physical form. In terms of computational evolution, there were two primary approaches covered (see slide below). One of these was AVIDA, which uses an encoded instruction set which can be used to perform a multitude of tasks. The other were variations of a genetic algorithm (GA), which uses a string of binary values as an analogy for DNA and chromosomes. Implicit to both approaches is that populations of these encodings are replicated and selected upon, which results in "evolved" solutions to problems.
Another approach to evolution was embodied by neurevolution, which simulates what goes on in the brain during learning or task performance. Rather than using a traditional GA, the neuroevolution approach utilizes a modified neural network (NN) architecture, as the focus of the model is neural activity rather than heredity. The Evolutionary Complexity group (EPLEX) -- University of Central Florida and the Neural Networks research group -- University of Texas are major sites of this research. The slide below shows the two main approaches (CPPN and HyperNEAT).
There were a number of talks on experimental evolution. There was an excellent conference on this topic held at the University of Michigan (ECSS 2010) in 2010. The slide below provides an overview of the Long-term Evolution Project run by Rich Lenski at Michigan State. Aside from paper and posters on that project, there was a presentation by Betul Kacar (Georgia Tech) on knocking in ancestral forms of key genes to create a "sub-optimal" bacterial phenotype. These populations would then evolve a means to compensate for this loss, which is experimentally tractable way to look at the evolvability of gene networks.
Below is a sampling of "emerging work" from the paper and poster talks. I chose these (see slide below) as they were the most intriguing to me. These four choices also demonstrate the broad diversity of topics covered.
There were upwards of 50 posters presented in a single session. The range of topics are listed below. As an aside, there were many different type of layout used, ranging from traditional layouts (text, data, conclusions) to more avant-garde approaches (non-sequential images with minimal text).
That's all for this year (which turned out to be quite a lot). Looking forward to Artificial Life XIV, which will likely be held in Europe (as the location alternates between Europe and North America). And keep on evolving....











ALife 14 will be in New York City in two years (it was decided at the meeting). ECAL 12 will be in Sicily
ReplyDeletehttp://www.dmi.unict.it/ecal2013/